 Cell-ACDC
Cell-ACDC
A GUI-based Python framework for segmentation, tracking, cell cycle annotations and quantification of microscopy data
 Source code on GitHub
Source code on GitHub
Written in Python 3 by Francesco Padovani and Benedikt Mairhoermann.
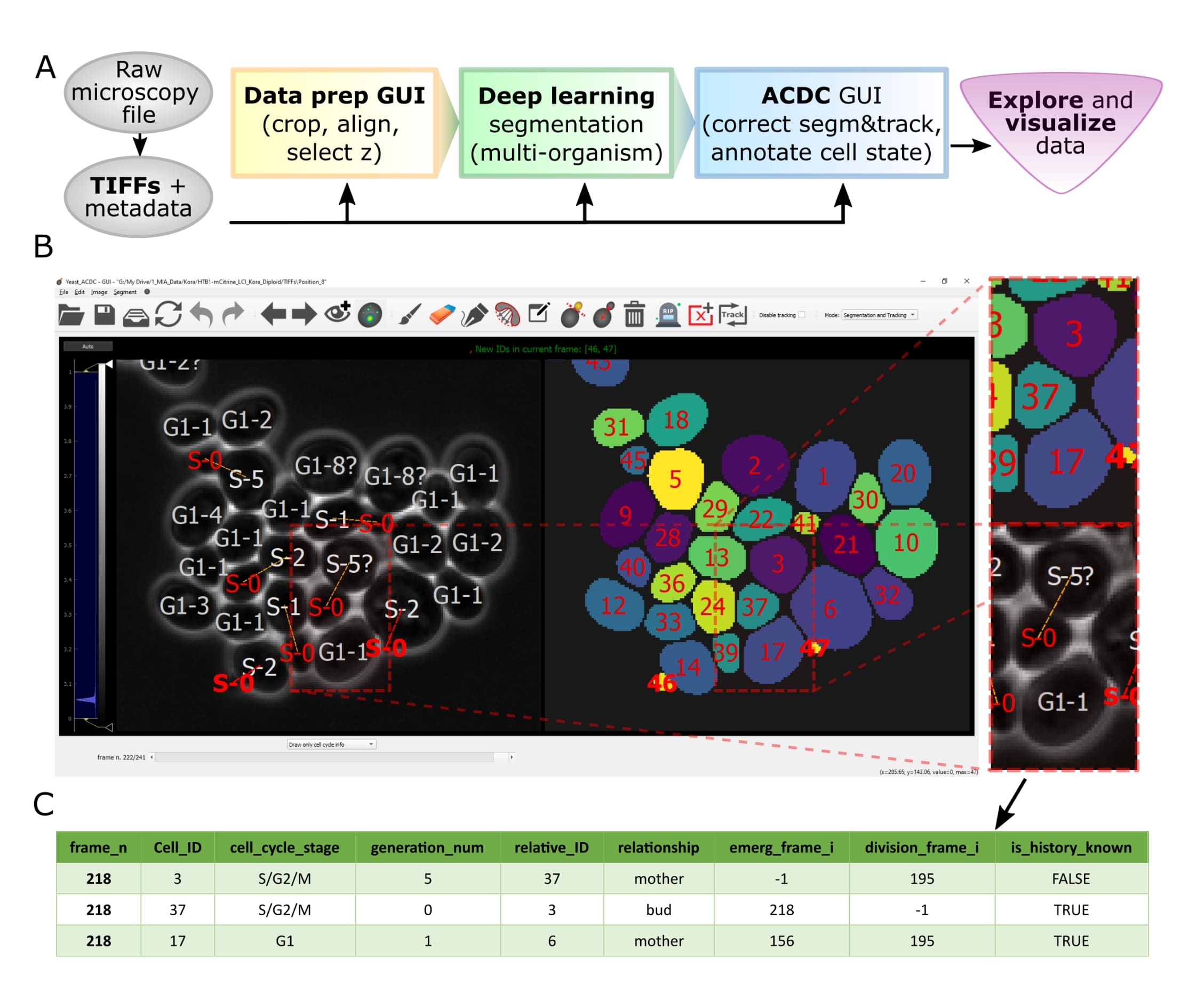
Overview of pipeline and GUI
Contents
- Overview
- Citation
- Installation
- Folder structure
- From 0 to Cell-ACDC mastery: A complete guide
- Create Data Structure with ImageJ/Fiji macros
- How to contribute to Cell-ACDC
- GUI tools
- Models for automatic segmentation and tracking
- Scripts to correct shifts in bidirectional scanning
- Cell-ACDC output data
- Troubleshooting
- Versions
- Resources





